Advancing epidemiology through the use of computation and digital tools.
Infectious diseases continue to pose substantial risks for human and animal populations globally contributing to societal disruptions and morbidity and mortality. Global processes such as increased urbanization, human mobility, and climate change together will continue to accelerate the speed and impact of infectious diseases. In order to make advances towards mitigating the impact from infectious diseases and improve global health our team develops computational tools and analyses that integrate novel digital and genomic data sources.
Moritz Kraemer is a Professor of Epidemiology & Data Science at the Department of Biology and Pandemic Sciences Institute, where he specialises in global infectious disease research. He is also a co-director on the Oxford Martin Programmes on Pandemic Genomics and Digital Pandemic Preparedness.
His research primarily focussed on global and equitable disease surveillance and how climatic, behavioural and evolutionary factors interact in shaping the epidemiology of diseases. He has a particular interest in how human behavioural patterns predict the risk of disease spread and how this knowledge can be used to inform mitigation strategies. Moritz's research has not only advanced academic knowledge but also informed public health policy decisions at national and international levels.
Moritz has received recognition for his contributions to how computational technologies like AI and machine learning can be used to advance health research (multiple Google AI faculty awards). He leads a global data science initiative, Global.health, that aims to advance the response to infectious diseases through the responsible use of open-source analysis software and sharing of epidemiological, clinical, genomic, and spatial data.
Before joining the University of Oxford, Moritz completed a fellowship at Harvard Medical School in Biomedical Informatics, where he focused on digital epidemiology. He holds a degree in statistical epidemiology from the University of Oxford.

Team
Peter J. Braam Early Career Research Fellow
How demographics and socio-economic factors shape infectious disease dynamics.
Data Scientist
Research Fellow
How evolutionary and behavioural factors interact in driving climate sensitive diseases.
DPhil Student
How multiple data types inform spatio-temporal dynamics of infectious diseases.
Postdoctoral Researcher
How network structure, human mobility and disease surveillance jointly shape inferences about key epidemiological parameters.
DPhil Student
How AI and computational analyses can be used to improve disease surveillance and public health decision making.
DPhil Student
I have a particular interest in modelling climate-sensitive, vector-borne diseases such as dengue fever.
Software Engineer
My focus is the development of scalable and locally deployable software to enable robust infectious disease analysis.
Data Scientist
I am interested in developing and deploying tools for enhanced data preparedness for pandemics.
Software Engineer
I built software to improve data preparedness, integration, and investigation during and ahead of epidemics and pandemics.
Visiting DPhil student
I am interested in arbovirus ecology and epidemiology in Thailand.
DPhil student
I work on the impact of human mobility and socio-economic factors in driving respiratory infections in East Africa.
- Dr. Houriiyah Tegally, Head of Data Science, CERI, Stellenbosch University
- Dr. Andrew Rambaut, Professor of Molecular Evolution, Edinburgh University
- Dr. Oliver Pybus, Professor of Evolution & Infectious Diseases, University of Oxford
- Dr. Marc Suchard, Professor of Biomathematics and Human Genetics, UCLA
- Dr. Christl Donnelly, Professor of Applied Statistics, University of Oxford
- Dr. Samuel Scarpino, Director of AI + Life Sciences, Northeastern University and Santa Fe Institute
- Dr. John Brownstein, Professor of Pediatrics and Biomedical Informatics, Harvard Medical School
- Dr. Samir Bhatt, Professor of Machine Learning & Epidemiology, Copenhagen University
- Dr. Simon Cauchemez, Head of Mathematical Modelling, Institut Pasteur
- Dr. George Githinji, Lead for Bioinformatics, KEMRI-Wellcome
- Dr. Nuno Faria, Professor of Virus Genomic Epidemiology, Imperial College London
- Dr. Anne Cori, Reader in Infectious Disease Modelling, Imperial College London
- Dr. Seth Flaxman, Associate Professor of Computer Science, University of Oxford
- Dr. Zoe Willis, Postdoctoral Fellow, 2020, now Scientist at the Office of National Statistics
- Dr. Thomas Rawsom, Postdoctoral Fellow 2020, Research Associate at Imperial College
- Graham Lee, Senior Software Engineer 2020-2021, Now Software Architect at Apple
- Gal Wachtel, Data Scientist 2021-2022, Now Senior Product Manager at ImmuneAI
- Dr. Anya Lindstroem Battle, Data Scientist 2020-2021, Now Postdoctoral Fellow at the University of Oxford
- Dr. Alexander Zarebski; Oxford Martin School Fellow 2018-2022, Now Lecturer at the University of Melbourne
- Zhiyuan Chen, Visiting Phd Student 2023-2024 from Fudan University
- Rosario Evans Pena; Data Scientist 2021-2024, Now Health Data Science DPhil student at the University of Oxford
Research
AI & Data Science
Through the collection and availability of large digital data sets and aided by fast and massively parallel computing hardware provide an opportunity for investigating infectious disease dynamics at unprecedented scales. For example, data extracted from satellite imagery, digital contact tracing, or mobile phones have a key role to play in understanding the emergence, distribution, and spread of infectious pathogens.
There remain, however, critical questions with respect to the reliability, ethics, and responsible use of digital data and algorithms to understand the health of populations. Our work on AI and digital tools is centered around the following themes:
- How can digitally collected data about human movements be integrated into disease models to predict infectious disease spread?
- How can AI be used to improve disease surveillance?
- How can the translation of scientific insights into public health policy be enhanced through the use of AI?
We also build digital tools for pandemic preparedness that are open-source and used by national and international governments worldwide.
We work with colleagues around the world in public health and academia through various funded projects on the above topics.
Moritz U.G. Kraemer et al. Artificial intelligence for modelling infectous disease epidemics. Nature 638, 623-635 (2025)
Moritz U.G. Kraemer et al. The effect of human mobility and control measures on the COVID-19 epidemic in China. Science 368,493-497 (2020)
Joseph L.-H. Tsui et al. Toward optimal disease surveillance with graph-based active learning. Proceedings of the National Academy of Sciences (PNAS) Vol. 121 | No. 52, e2412424121 (2024)
Oxford Martin School Programme on Digital Pandemic Preparedness Oxford Martin School
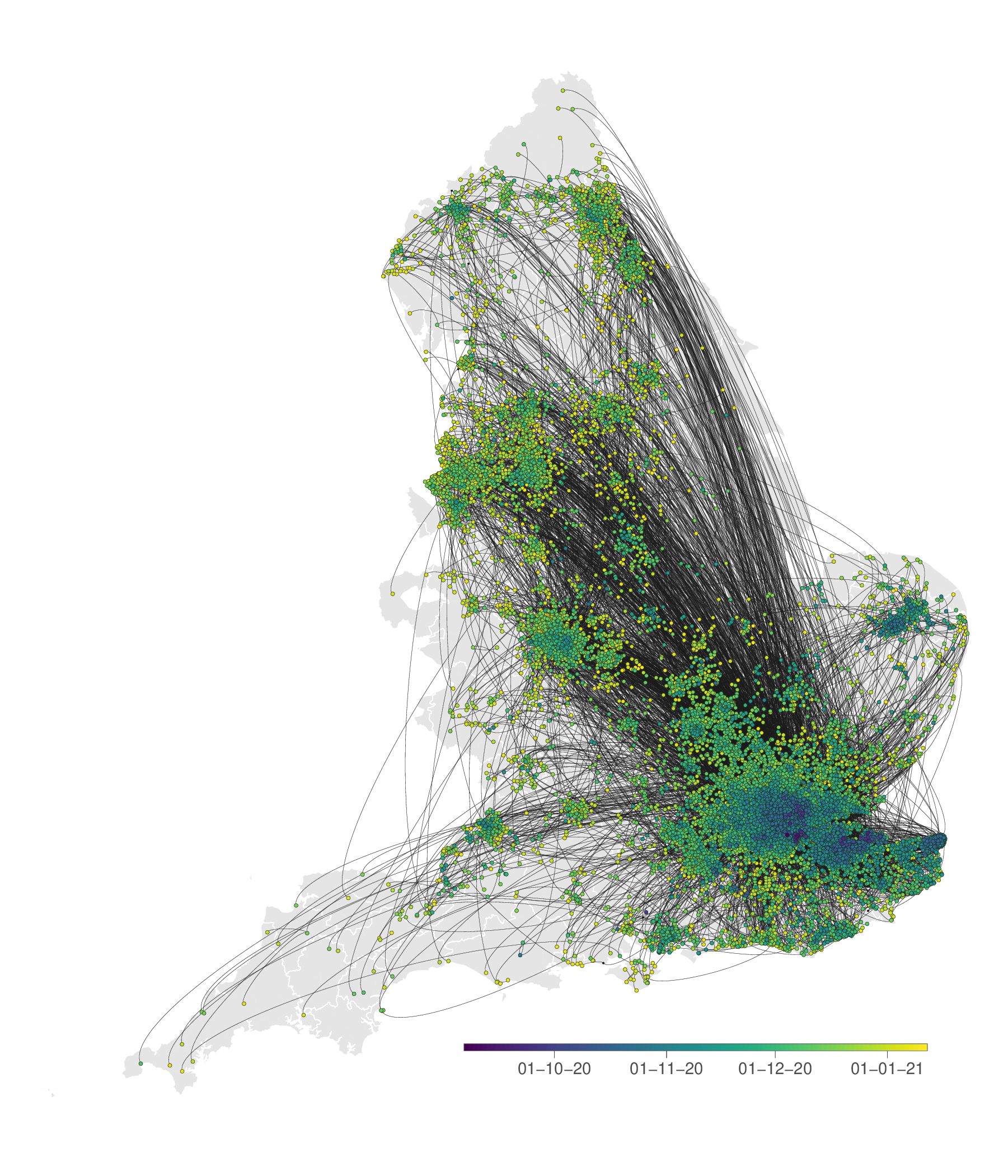
Genomics & Phylodynamics
New sequencing technologies and global capacity to generate pathogen genomes mean that entirely new aspects of pathogen transmission can be revealed.
Our research programme on pathogen genomics and epidemiology brings together mathematical epidemiology, pathogen phylodynamics, and human geography to build a new body of theory capable of co-analysing these distinct sources of information to identify the sources, maintenance, and spread of pathogens in populations. Our current research topics include:
- We develop methods for the analysis of large, pandemic scale genomic datasets
- Which sampling frameworks are optimal for identifying threats from pathogens and to reconstruct outbreaks when sampling is limited?
- How can pathogen phylodynamics become more predictive?
Much of this work is performed in close collaboration with Prof. Oliver Pybus.
Moritz U.G. Kraemer et al. Spatiotemporal invasion dynamics of SARS-CoV-2 lineage B.1.1.7 emergence. Science 373, 889-895 (2021)
Houriiyah Tegally et al. Dispersal patterns and influence of air travel during the global expansion of SARS-CoV-2 variants of concern. Cell 186, 3277-3290 (2023)
Zhiyuan Chen et al. COVID-19 pandemic interventions reshaped the global dispersal of seasonal influenza viruses. Science 386 (2024)
Oxford Martin School Programme on Pandemic Genomics Oxford Martin School
Climate & Infectious Diseases
Climate change is expected to increase the burden of infectious diseases around the world. A recent study found that over half of all known human infectious diseases can be made worse by climate changes. The major pathways how climate impacts infectious diseases are:
- Climate change creates more opportunities for viruses to be shared between species that were previously isolated, leading to more zoonotic transmission events and potential spillovers into human populations.
- Climate change expands the environmental suitability of disease vectors and climate-sensitive diseases.
- Higher temperatures due to climate change can accelerate pathogen transmission.
- Extreme weather events can increase the burden of infectious diseases by disrupting health systems, displacing populations, and accelerating infectious disease transmission in crowded settings.
We use statistical and mechanistic models to advance our broad understanding of how spatial heterogeneities impact transmission dynamics of infectious diseases. Much of our ongoing work in this area uses large-scale climate, socio-economic, and disease data to create better evidence on the links between climate and infectious diseases. To do so we work with partners in South Africa, Vietnam, Fiji, and the Americas.
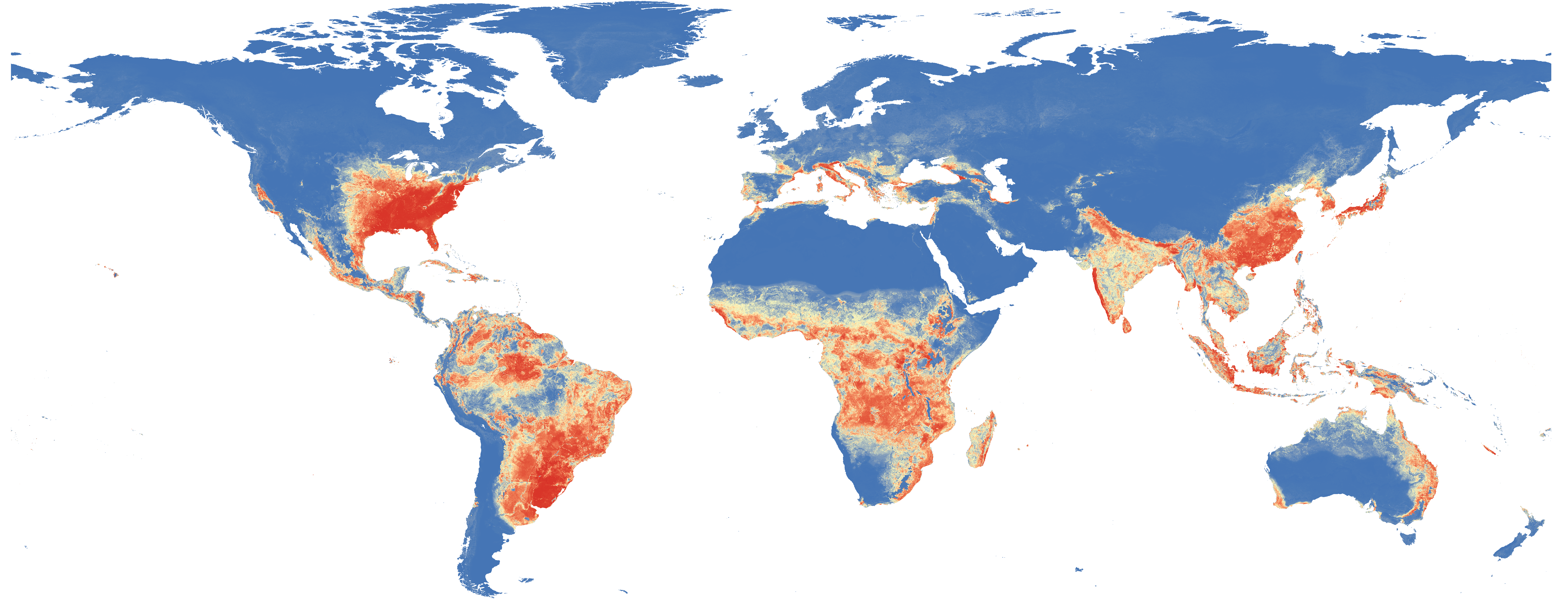
Moritz U.G. Kraemer et al. Past and future spread of the arbovirus vectors Aedes aegypti and Aedes albopictus.. Nature Microbiology 4, 854-863 (2019)
Tsui et al. Impacts of climate change-related human migration on infectious diseases. Nature Climate Change 14, 793-802 (2024)
Project on climate and infectious diseases Medical Research Foundation
Eco-Epidemiological Intelligence for Early Warning and response to Mosquito-borne disease risk in Endemic and Emergence Settings European Union & UK Research and Innovation


Open Source
Pandemic and epidemic response relies heavily on the availability of timely and accurate epidemiological, genomic, and clinical data. These data are used to answer key questions about the trajectory of an epidemic, its origins, and severity, informing interventions, treatments, and vaccination distributions. Global.health is a global consortium founded in 2020 to tackle challenges in data sharing, routed in principles of equitable and responsible global collaboration. You can find out more at:

Transmission of arboviruses such as Zika, Dengue, Chikungunya, and Yellow fever have increased substantially over the last decades causing significant concern for public health. We produced maps and data for the distribution of their disease vectors and made both available openly under permissive licenses.
Our lab builds sofware that make the integration, harmonization and analysis of data possible. To do so we publish code and documentation through our GitHub repositories.
Join Us
We train enthusiastic, creative, and diverse researchers from around the world at all career stages. Joining our team means becoming part of one of the best and innovate research Universities in the world. Contributions to the lab and University are valued beyond the institution and have the potential to generate differences in global health.
DPhil students
We accept DPhil students through several departments and programmes at the University of Oxford, including DPhil in Biology, DPhil in Population Health, Intelligent Earth UKRI CDT in AI for the Environment. You may apply to more than one programme. Prospective DPhil students with an excellent academic record should email a few paragraphs outlining their background and prior experience, motivation, and research interests in our lab. They might also be interested in working with others at Oxford and students are often co-advised by 2-3 academics. You can find out more about graduate admission here and graduate funding here.
Postdocs and Research Fellows
We regularly advertise new postdoctoral research positions on this website and through the University of Oxford Vacancies page. Candidates with a strong research and analytical background, excellent publication record, and demonstrated ability to perform independent research should email me (moritz
Undergraduate researchers
Undergraduate researchers from the University of Oxford can work with us on their final year research projects or do internships over the summer (as long as it does not interfere with the students coursework). Interested students should get in touch describing their interest.